👋🏼 Hey there, I’m Sohith!
I’m a final-year Bioinformatics undergrad at Saveetha School of Engineering, SIMATS University, Chennai, fueled by a passion for blending neuroscience, structural biology, and AI to solve real-world healthcare challenges.
What I’m Working On
Right now, I’m a student researcher at the University of Colorado Colorado Springs (with Ioncure), building AI tools using NLP, Computer Vision, and Machine Learning to raise awareness about neurodegeneration.
I also dive into computational drug discovery, AI-driven diagnostics, and molecular modeling—pretty exciting stuff!
My Journey So Far
Over the past year, I’ve been lucky to work with some amazing teams across Asia:
- Chang Gung University, Taiwan: Explored molecular docking to study drug-target interactions.
- KEK, Japan: Worked on X-ray crystallography and Cryo-EM at the Structural Biology Research Center, and wrote English technical manuals for future interns.
- National Dong Hwa University, Taiwan: As a TEEP@Asia+ Scholar, I built a machine learning pipeline for TB/NTM classification using mass spectrometry data, hitting a 0.98 AUROC and 0.94 F1-score.
I’ve also published a few papers, filed patents, and mentored STEM students to make science more approachable. Through books, podcasts, and blogs, I love sharing knowledge and sparking curiosity.
Beyond the Lab
When I’m not geeking out over science, you’ll find me exploring new places—sipping coffee in cozy Taiwanese cafés or savoring ramen in Tokyo’s lively streets.
These adventures often inspire my next research idea. I’m excited to bring this curiosity into my PhD journey.
Let’s Connect!
If you’re into neuroscience, AI in healthcare, or just want to chat research over coffee, I’d love to hear from you.
Let’s make science happen together!
Research Experience
Current Experience
Undergraduate Student Researcher, University of Colorado Colorado Springs (UCCS), Colorado Springs, United States of America (Remote)
DEC’ 24 - Present
Working in collaboration with the University of Colorado and Ioncure to develop and apply AI-driven methods for evaluating neurodegeneration awareness. Leveraging Natural Language Processing (NLP), Computer Vision, Machine Learning, and advanced data science techniques to analyze diverse data sources.

 Research Assistant, Manipal University College, Melaka, Malaysia (Hybrid)
Research Assistant, Manipal University College, Melaka, Malaysia (Hybrid)
MAY’ 24 - Present
Analyzed Solanum nigrum extract for antibacterial compounds using GC-MS data, successfully identifying 50 bioactive components under the guidance of Dr. Sugapriya. This research has laid the groundwork for novel antimicrobial investigations using plant-based compounds. Conducted in silico docking studies on M. tuberculosis MurE ligase using AutoDock Vina to identify potential anti-TB agents from S. nigrum extract. Currently, I am assisting with writing research papers, contributing to book chapters, and supporting various ongoing projects in the field of antimicrobial research.
Previous Experience


JULY' 25 - AUG' 25
Worked at Structural Biology Research Center (SBRC), Institute of Materials Structure Science, High Energy Accelerator Research Organization (KEK), under Toshio Moriya-sensei & Miki Senda-sensei. Actively engaged in structural biology research focusing on X-ray crystallography refinement and Cryo-Electron Microscopy (Cryo-EM) analysis of protein structures. Assisted in the creation of beginner-friendly English manuals and technical documentation to support future international interns and early-stage researchers at KEK, covering common workflows and troubleshooting in macromolecular crystallography.

Lab members of SBRC group during our dinner pizza party 🍕🎉

 Research Intern, National Dong Hwa University, Hualien, Taiwan (On-site)
Research Intern, National Dong Hwa University, Hualien, Taiwan (On-site)
JAN’ 25 - JUN’ 25
Operated MALDI-TOF mass spectrometer at Biophysics Mass Spectrometry Lab, NDHU for TB and NTM sample analysis under the supervision of Dr. Wing Peng-Ping and developed ML models for tuberculosis classification using acquired spectral data and performing statistical analysis and feature selection to identify significant m/z peaks, coupled with spectral data visualization for enhanced interpretability.

Lab members of BMS group at Liyu Lake after a trek 🌿
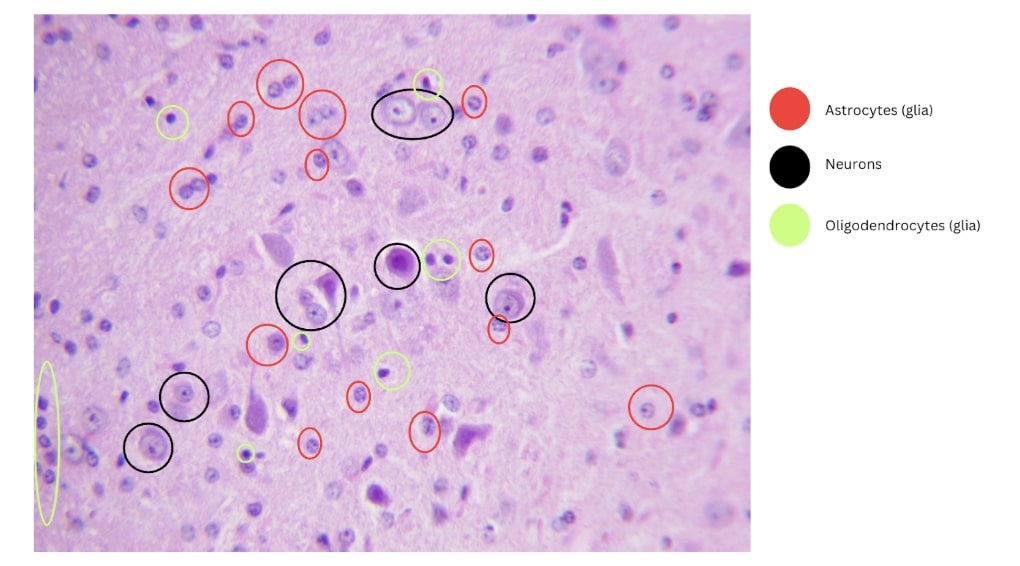 Research Intern, Joint Institute for Nuclear Research, Moscow, Russia (Remote)
Research Intern, Joint Institute for Nuclear Research, Moscow, Russia (Remote)
MAR’ 25 - APR’ 25
During this internship, I focused on histological analysis of the central nervous system post-irradiation, with two key objectives. First, I performed manual classification of neurons and glial cells using ImageJ’s Cell Counter plugin. I mapped anatomical brain regions with guidance from the Allen Brain Atlas and classical histology references. Second, I quantified apoptotic cell death by analyzing TUNEL-stained sections to detect and compare TUNEL-positive cells between control and irradiated samples. This integrated approach combined traditional neurohistology with digital image analysis to enhance the accuracy and efficiency of CNS pathology assessment. Report Link
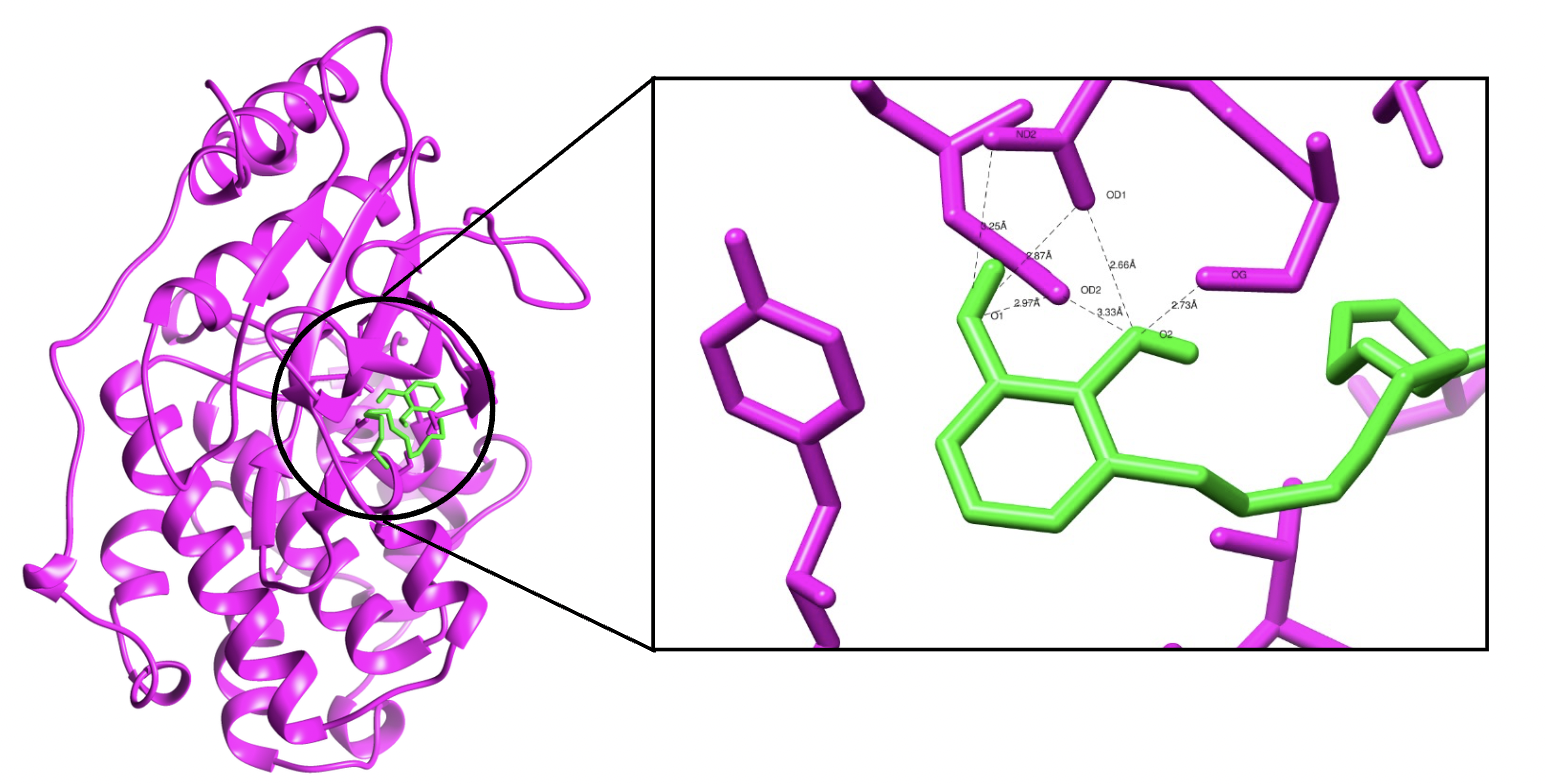 Research Intern, Genomac Hub, Ogbomosho, Nigeria (Remote)
Research Intern, Genomac Hub, Ogbomosho, Nigeria (Remote)
SEP’ 24 - NOV’ 24
I worked on selecting peptides with potential anti-cancer activity by analyzing key biochemical properties such as hydrophobicity, net charge, stability, half-life, and Boman index. These properties help determine a peptide’s ability to interact with cancer cell membranes, stability in biological environments, and target specificity. Using correlation heat maps and PCA analysis, our team identified 28 bacterial peptides, 18 probiotic bacterial peptides, 6 fungal peptides, and 4 metagenomic peptides. My specific focus was on peptides from the soil bacterium Streptomyces parvus, contributing to the overall findings of potential anti-cancer candidates. Graduation Ceremony
Undergraduate Research Student, Saveetha School of Engineering, Chennai, India (On-site)
JAN’ 23 - MAR’ 24
Under the supervision of Dr. Kannan, I conducted data preprocessing and exploratory data analysis (EDA) on the ChEMBL dataset for Acute Myeloid Leukemia’s drug discovery using pandas and matplotlib/seaborn. I implemented and compared various machine learning models (Random Forest, SVM, XGBoost), achieving an accuracy of 82% with Random Forest for predicting potential drug candidates. Additionally, I performed molecular docking analysis using AutoDock Vina, applied Lipinski’s Rule of Five with RDKit, and executed molecular dynamics simulations using GROMACS to validate and analyze protein-ligand interactions of promising Alzheimer’s drug candidates.
